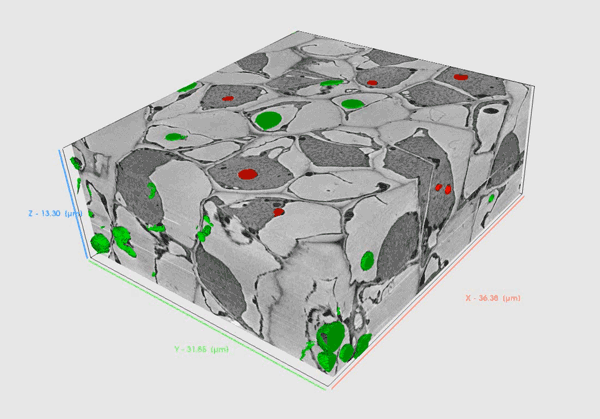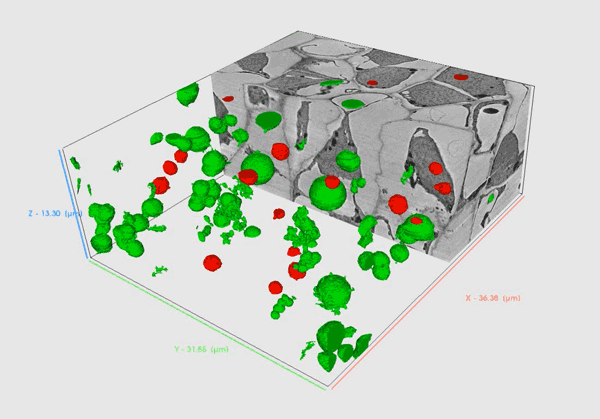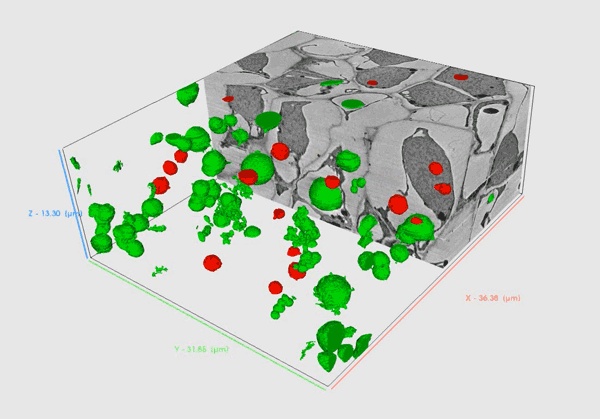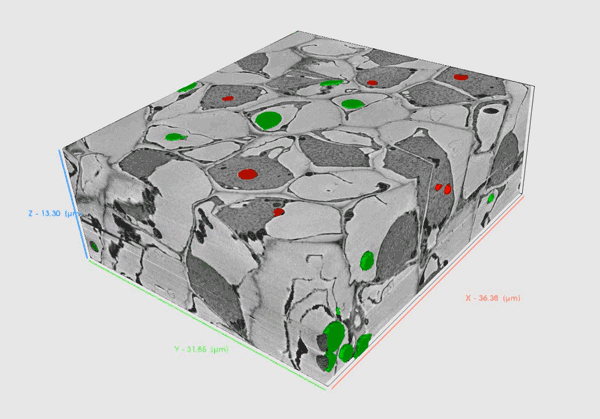
Volume SEM methods provide insights into biological material in high resolution that allows researchers to comprehensively understand the internal structure and functioning of biological systems in 3D.
The 2D imaging capability of almost any SEM can be expanded to a 3rd dimension with specialized software or hardware. Method of choice depends on the priority and character of the sample. Samples that have been sectioned beforehand, collected on a substrate as an array of serial sections (Array Tomography) can be automatically scanned with a SEM to produce a 3D stack of images. This method allows repeated scanning and the ability to go back to additional regions of interest at any time. On the other hand, a SEM equipped with a special in-chamber microtome is able to analyze volumes of exceptional dimensions in the field electron microscopy – blocks of almost one cubic millimeter can be examined.This allows us to understand the complex structural organization of cells, tissues, organs and organisms, and relationships over a large distance, such as animal connectomes.
Microscope flexibility, throughput and ease of use are of utmost importance for instruments in core facilities and imaging centers. Therefore, TESCAN volume SEMs are designed to be fully operational after only a short training session. This is possible with our modular Essence™ GUI that is configurable and predefined for specific applications. Quality of results is assured by streamlining the process from easy acquisition setup through to data processing, machine-learning assisted segmentation and visualization in a single, unified TESCAN software environment.
In-chamber microtome slicing & imaging (SBF-SEM)
Serial Block Face – SEM (SBF-SEM) utilizes an in-chamber microtome to allow large volume analysis by in-situ sectioning and imaging of samples. Samples are usually resin-embedded blocks of cells, tissues or small organisms. SBF-SEM is used for larger volumes as it enables slicing of areas as wide as 1mm enabling multiple regions of interest to be acquired during the run. Precise microtome slicing through the sample together with an efficient detection system allows for the acquisition of large datasets with a modest Z resolution in a reasonable time with swift reconfiguration back to a normal SEM regime when the acquisition is complete.
In Array Tomography, a resin-embedded tissue specimen is cut into ultrathin sections which are then bonded to a silicon wafer outside the SEM chamber. The resulting array is scanned automatically with a dedicated software module. This approach is especially beneficial when the sample needs to be preserved for future reference or for immuno-labelling of sections.
This method does not require any specialized hardware to be installed on the SEM. Streamlined software guides the user through the acquisition setting in a simple and intuitive manner and also offers a straightforward way of processing the resulting data to create 3D visualizations.
-
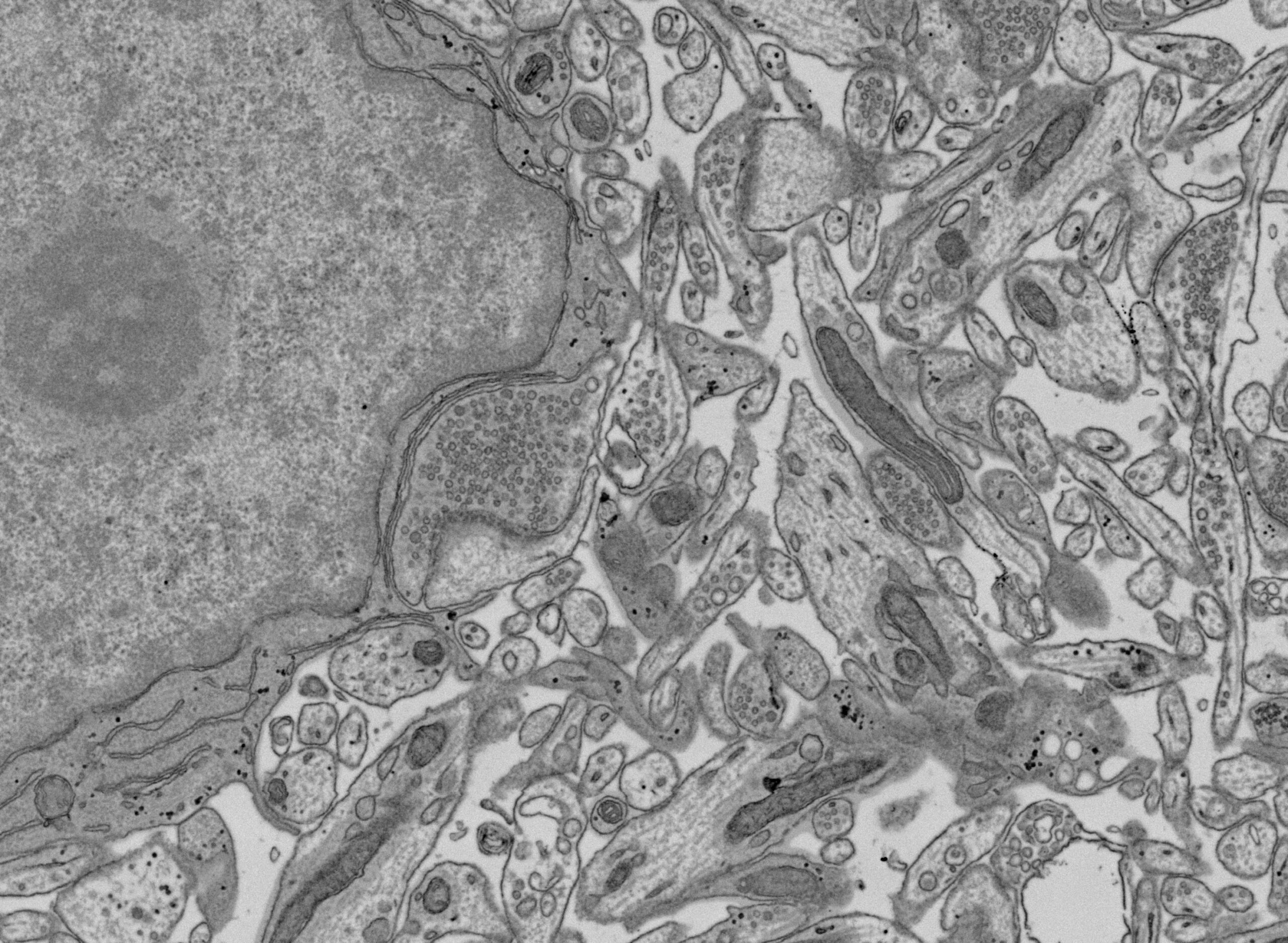
- Thin section of mouse brain on silicon wafer imaged with BSE detector, inverted contrast. Image courtesy: Dr. Rudolph Reimer, Leibniz Institute of Virology.
-
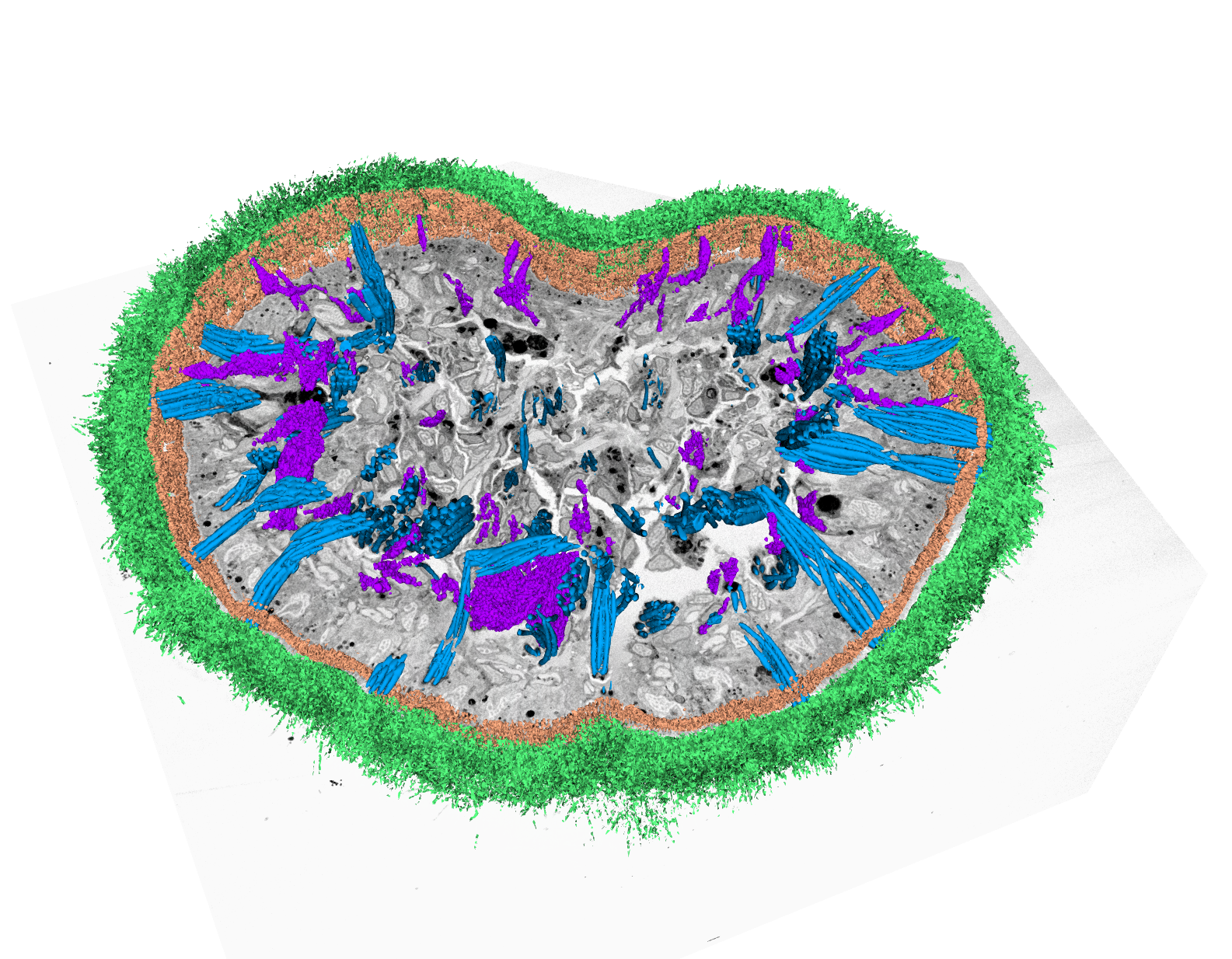
- Flatworm Macrostomum lignano, visualization of volume dataset acquired by SBF-SEM with segmented cilia (green) and rhabdites (blue)